Cell Line Identification
Methods for Authentication of Human Cell Cultures and Tissues
Childhood Cancer Repository, powered by Alex's Lemonade Stand Foundation
Pediatric Cancer Center, Texas Tech University Health Sciences Center School of Medicine,
Lubbock, TX 79430, U.S.A.
Introduction
Growth of cells in cultures or xenografts provides many opportunities for mislabeling
or cross-contamination in spite of best laboratory practice. Contamination can cause
mixtures with resulting outgrowth and subsequent replacement by the more aggressive,
dominant line. Therefore the genetic identities of cell cultures and xenografts should
be validated when initially established and verified periodically to insure the integrity
of research conducted with the lines (1). On November 28, 2007, the National Institutes
of Health issued a notice that in part made the following statements (2);
"We are convinced by the body of evidence developed by Dr. Nardone and his colleagues that misidentification of cell cultures is a serious problem. Grant applications that fail to employ such practices (cell culture authentication to prove true identity of the culture) would not be considered of the highest quality and such manuscripts would not fare well in the journal review process. We encourage all reviewers to consider these issues carefully in order to protect and promote the validity of the science we support."
The Texas Tech University Health Science Center (TTUHSC) School of Medicine (SOM) Pediatric Cancer Center Cell Culture Core makes available methods of molecular identification of cell lines and xenografts. After study of the various alternatives, the laboratory chose to implement the Applied Biosystems AmpF/STR® IdentifilerTM system for genetic identification of cell lines and xenografts. The IdentifilerTM kit was developed specifically for use in forensic laboratories throughout the world (3) as a standardized method for assessing short tandem repeat (STR) loci and software to reduce analyzed loci to a numeric "molecular fingerprint". This method is routinely employed in the Childhood Cancer Repository, powered by Alex's Lemonade Stand Foundation (4).
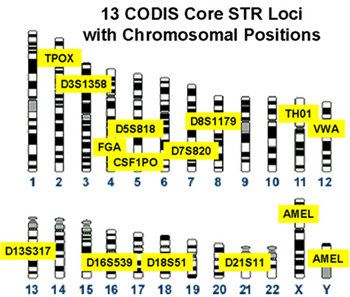
Figure 1. Loci designated for use in the Combined DNA Identification System (CODIS). These loci were selected by the Federal Bureau of Investigation (FBI) after years of research and collaboration with other laboratories. The CODIS database is maintained by the FBI and contains offender DNA profiles generated by Federal, State and local crime laboratories. Comparing these 13 required forensic loci to those used in the IdentifilerTM kit you will see that all loci from the FBI CODIS are included in the IdentifilerTM kit, and IdentifilerTM employs 2 additional loci (D2S1338 and D19S433) (Figure 2).
The IdentifilerTM kit is a human specific, multiplex PCR-based system testing 15 different short tandem repeat (STR) polymorphic loci. A 16th locus, intron 1 of the amelogenin gene, which is located on the X and Y chromosomes, provides gender identification. A 6 base pair deletion on the X chromosome allele of amelogenin allows electrophoretic distinction between DNA from X or Y chromosomes. Since females are X,X, only a single peak is observed when testing female DNA whereas males, which possess both X and Y chromosomes, exhibit two peaks with a standard amelogenin test (3).
The loci chosen have high heterozygosity, regular repeat units, distinguishable alleles and robust amplification (2). The primers used during PCR have fluorescent dyes attached and the resulting amplicons are laser-detected as fragments of different lengths during capillary electrophoresis. Then, proprietary software is used to establish profiles based upon the number of repeated sequences at each allele of each locus. The loci used are located on independently assorting chromosomes and the probability of this particular assay for identifying tissues from two different individuals as identical averages from 1 in 7.64 x 1017 to 1 in 2.76 x 1016, depending on the race (5).

Figure 1A. Example of short tandem repeat (STR) sequence of DNA (TCAT). This allele would type as a "6".
Materials and methods
DNA samples and STR profiling
Human cell culture material, tumors and whole blood samples are the most common samples
submitted to the COG Core Laboratory for STR typing. Presumed xenograft tumors from
mice are also submitted to determine if the tissue is of human or mouse origin since
this assay is human specific. Cell culture material is submitted as dry pellets consisting
of 1 x 106 to 5 x 106 cells and placed in the Core Laboratory -20°C freezer. Tumor samples are submitted
as approximately 25 mg of tissue pieces in microcentrifuge tubes and whole blood samples
are submitted as approximately 1 ml in microcentrifuge tubes. The contributor logs
the samples into the STR Samples Request book at the time of the submission.
DNA is isolated from the cell pellets or tissues with the Qiagen® DNeasy® kit and quantitation of the extracts is performed using a NanoDropTM spectrophotometer (Thermo Fisher Scientific Inc). DNA amplification and fragment fluorescent labeling is performed using the GeneAmp® PCR System 9700 thermal cycling instrument and the IdentifilerTM AmpF/STR® PCR kit. Approximately 0.125 ng/µl of purified DNA is used for amplification (6).
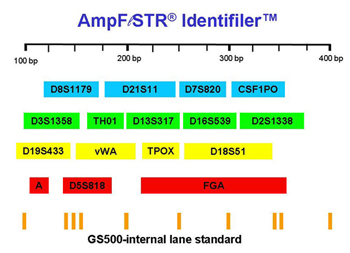
Figure 2. Loci amplified when using the IdentifilerTM kit.
Capillary electrophoresis and fragment detection is performed with a 3130xl Genetic Analyzer with Data Collection software v 3.0 (Applied Biosystems). GeneMapper ID® Version 3.2.1 (Applied Biosystems) software is used to assign fragment repeat numbers which becomes collectively the STR type for that individual (6).

Figure 3. Thermal cycler and ABI 3130xl Genetic Analyzer, 16 capillaries.
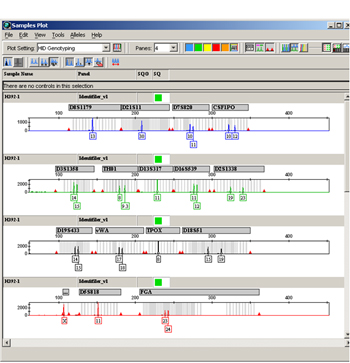
Figure 4. Example of GeneMapper ID® STR data.
Database Searches
The result of DNA STR typing done for the Childhood Cancer Repository and TTUHSC includes
the complete 16 loci set of the sample, which are entered into an MySQL database and
the results are searchable on the strdb.cccells.org website. This database currently contains ~ 2500 previous STR profiles from cell
cultures and tissues from the Childhood Cancer Repository, cell lines established
at TTUHSC and UT Southwestern Medical Center, and STR data from other repositories
(ATCC, DSMZ, JCRB, and Riken). Some of the data employ 8 loci profiles. The sample contributor is advised of any
matches by email. This database is employed internally by the Childhood Cancer Repository, powered by Alex's Lemonade Stand Foundation, and the TXCCR when establishing new lines or validating established cultures. The database is available
online (https://strdb.cccells.org/) for any investigator to use to identify a cell line or verify that a new line is
unique with respect to the database of known lines.
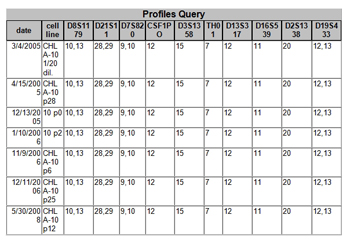
Figure 5. Example of matches obtained when a new sample profile was entered into the database
with ten loci shown. The 2008 cell line is the same as the cell line first typed in
2005. The "p" in the cell line name stands for the passage number.
Acknowledgements
This web page was based on a poster designed by Larry Starr, TTUHSC, Medical Photography, Lubbock, TX and Charles Linch, TTUHSC Pediatric Cancer Center.
References
- Newsfocus: Cases of mistaken identity. Science Vol 315, page 928-931, Feb 16, 2007
- http://grants.nih.gov/grants/guide/notice-files/NOT-OD-08-017.html
- www.cstl.nist.gov/strbase/
- Cabral DJ, Feaman HV, Sheard MA, Reynolds CP: Short tandem repeat genotyping using IdentifilerTM is a reproducible and cost-effective method for establishing and monitoring identity of pediatric cancer cell lines. Proc Amer Assoc Cancer Res 48:96, 2007.
- http://www3.appliedbiosystems.com/cms/groups/.../cms_041201.pdf
- Standard Operating Procedure, SOP 25: Short Tandem Repeat STR Profiling GP10.